The plummeting costs and rising availability of genetic sequencing are lowering the threshold for such testing while also magnifying the questions that swirl around individual results.
Do patients want to know their proclivities for disease? Should they be told so their family can be tested? Does knowing or not knowing produce greater anxiety?
Dan Roden, M.D., senior vice president for personalized medicine at Vanderbilt University Medical Center, is using large EHR datasets and new high-throughput cellular studies to answer these kinds of questions and put genetic testing on firmer ground.
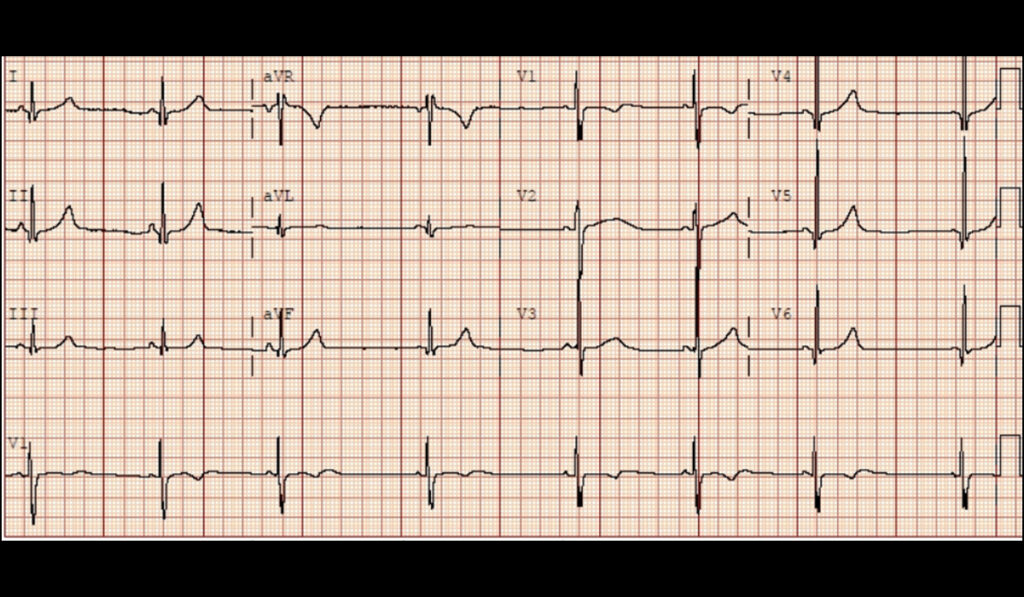
Roden is site co-principal investigator in the NIH’s Electronic Medical Records and Genomics (eMERGE) Network study, one the first studies to explore the frequency and clinical impact of rare genetic variants in a broad population. Through eMERGE-III, the network sequenced 109 Mendelian disease genes, including 10 associated with arrhythmias, in 21,846 individuals who had no indication for arrythmia-related genetic testing.
“The study highlighted the problem of variants of unknown significance, which constitute the largest proportion of genetic variants. Their numbers and potential diagnostic significance make them the new frontier for sequencing to test for arrhythmias,” Roden said.
Unknown Significance Stands Out
In the study, researchers found 120 participants with a pathogenic genetic arrhythmia condition. For these patients, the finding meant new treatments, lists of drugs to avoid, and further screening for family members.
The pathogenic variants were only the tip of the iceberg, however. In 1,838 patients, researchers found never-before-seen varients in the 10 genes associated with genetic arrhythmia. These were classified as variants of uncertain significance (VUS).
“The study highlighted the problem of variants of unknown significance. Their numbers and potential diagnostic significance make them the new frontier for sequencing to test for arrhythmias.”
In 50 patients, these unknown variants were also associated with arrhythmia codes in electronic medical record systems, so Roden’s group performed cellular studies to look at their function. Those studies led to reclassifying the variants as pathogenic in eight cases and benign in three.
“With a VUS, it is unknown whether it does nothing to the function of the protein that the gene encodes, completely destroys protein function, or alters biology in some other way,” Roden said.
He said it is important to try to classify these variants to avoid having entire families screened unnecessarily for a benign variant.
“There’s genetic sequencing everywhere, and our challenge is to ensure that it’s used right,” Roden said. “We need to make sure patients don’t get crippled by anxiety, make sure testing that can save lives is done, and that the health-care system doesn’t get bankrupted by unnecessary testing after genetic testing.”
Developing Variant Effect Maps
Roden and colleagues have been awarded a large NIH grant to develop assays that can determine the function of all possible variants within genes selected for study.
“We will look at function in many systems, from cells that express individual proteins to pluripotent stem cells to yeast,” Roden said.
“We subscribe to the vision that the term ‘VUS’ needs to become obsolete as we are increasingly able to assess pathogenicity.”
The group plans to study variants in major genes known to be involved in arrhythmias, cardiomyopathies and cholesterol disorders. For each gene, they will first make a pool of thousands of possible single variants. The assays that the group develops will then be used to analyze the pool and identify variants that disrupt protein function and those that don’t and can be designated as benign.
“To display the results, we generate what we call a variant effect map which is color coded to show which variants are pathogenic and which aren’t,” Roden said.
In the Clinic
“We subscribe to the vision that the term ‘VUS’ needs to become obsolete as we are increasingly able to assess pathogenicity,” Roden said. “That will not only help us understand how these proteins work, but also directly help our patients.”
In one case, a rare variant was found in an 11-year-old child who had been resuscitated from a cardiac arrest caused by an abnormal rhythm. This variant was initially designated as a VUS.
“Using our map, we were able to tell the treating doctor that the variant was pathogenic,” Roden said. “That helped explain why the patient had a cardiac arrest and prompted genetic screening of family members.”
Another application of this approach is in patients with non-ischemic cardiomyopathy.
“This can be a particularly nasty disease, and variants in some cardiomyopathy genes can send patients to defibrillators or a transplant list,” he said. “When we find a VUS in one of those genes, the variant effect maps can help make critical patient-care decisions.”